Document Type : Regular Article
Authors
1
Department of Chemistry, Faculty of Sciences Dhar el Mahraz, Sidi Mohammed University, Fez, Morocco.
2
2Laboratory of Processes, Materials, and Environment (LPME), Faculty of Science and Technology, Sidi Mohamed Ben Abdellah University, Fez
3
Engineering Laboratory of Organometallic, Molecular Materials, and Environment, Sidi Mohamed Ben Abdellah University, Faculty of Sciences, Fez, Morocco.
4
MCNS Laboratory, Faculty of Science, University Moulay Ismail, Meknes
5
1Engineering Laboratory of Organometallic, Molecular Materials, and Environment, Sidi Mohamed Ben Abdellah University, Faculty of Sciences, Fez,
6
Department of Chemistry, Faculty of Sciences Dhar el Mahraz, Sidi Mohammed Ben Abdellah University, Fez, Morocco.
7
EST Khenifra, Sultan Moulay Sliman University
10.22036/pcr.2022.370609.2241
Abstract
Cyclin-dependent kinase 2 (CDK2) has appeared as a promising healing goal for anticancer treatments. Thus, this work seeks to predict new nominee drugs against cancer. In the present paper, a three-dimensional quantitative structure-activity relationship (3D-QSAR), molecular docking, and molecular dynamic simulation (MD) were put in to search the binding between CDK2 and Styrylquinoline inhibitor to design novel anticancer agents. The best 3D-QSAR models were performed via Comparative Molecular Field Analysis (CoMFA) and Comparative Molecular Similarity Indices Analysis (CoMSIA) with elevated values of Q2of 0.580, and 0.68, as well as values of R2= 0.90 and 0.91, sequentially. The forecasting ability of the models was tested by Y-randomization and external validation employing a test set of 9 molecules with a predicted determination coefficient Rtest2 of 0.99 and 0.94 for CoMFA and CoMSIA respectively. The molecular docking approach with Sybyl X.2 is conducted in this study to enhance the analysis taken out from CoMFA and CoMSIA contour maps and to afford an in silico search for the best appropriate method of inhibitor interaction within its enzyme .Furthermore, the dynamic performance and strength of the high-activity molecules were tested by performing MD simulations. This study provides leadership in to design of new potent molecules.
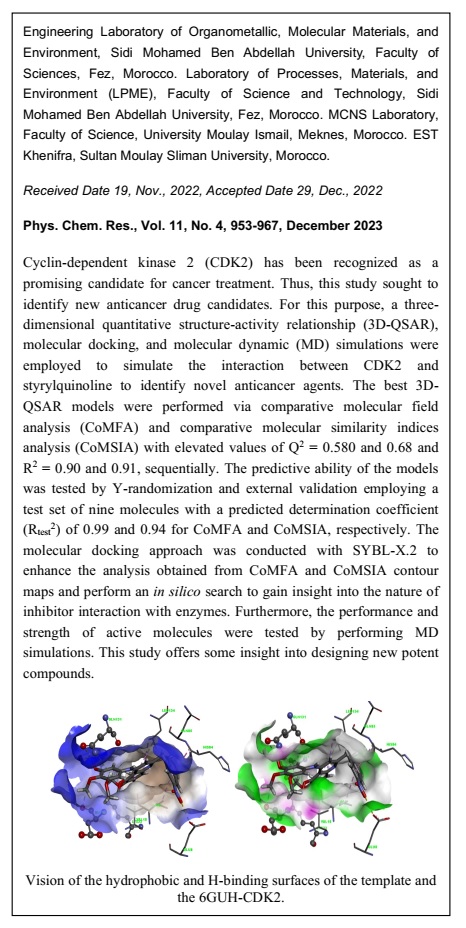
Graphical Abstract
Keywords

